Plotting method for objects of class fkf. This function
provides tools for graphical analysis of the Kalman filter output:
Visualization of the state vector, QQ-plot of the individual
residuals, QQ-plot of the Mahalanobis distance, auto- as well as
crosscorrelation function of the residuals.
Arguments
- x
The output of
fkf.- type
A string stating what shall be plotted (see Details).
- CI
The confidence interval in case
type == "state". SetCItoNAif no confidence interval shall be plotted.- at.idx
An vector giving the indexes of the predicted state variables which shall be plotted if
type == "state".- att.idx
An vector giving the indexes of the filtered state variables which shall be plotted if
type == "state".- ...
Value
Invisibly returns an list with components:
distance | The Mahalanobis distance of the residuals as a vector of length \(n\). |
std.resid | The standardized residuals as an \(d \times n\)-matrix. It should hold that \(std.resid_{ij} \; iid \sim N_d(0, I)\), |
where \(d\) denotes the dimension of the data and \(n\) the number of observations.
Details
The argument type states what shall be plotted. type
must partially match one of the following:
stateThe state variables are plotted. By the arguments
at.idxandatt.idx, the user can specify which of the predicted (\(a_{t}\)) and filtered (\(a_{t|t}\)) state variables will be drawn.resid.qqDraws a QQ-plot for each residual-series in
vt.qqchisqA Chi-Squared QQ-plot will be drawn to graphically test for multivariate normality of the residuals based on the Mahalanobis distance.
acfCreates a pairs plot with the autocorrelation function (
acf) on the diagonal panels and the crosscorrelation function (ccf) of the residuals on the off-diagnoal panels.
usage
plot(x, type = c("state", "resid.qq", "qqchisq", "acf"),
CI = 0.95, at.idx = 1:nrow(x$at), att.idx = 1:nrow(x$att), ...)
See also
Examples
## <--------------------------------------------------------------------------->
## Example: Local level model for the treering data
## <--------------------------------------------------------------------------->
## Transition equation:
## alpha[t+1] = alpha[t] + eta[t], eta[t] ~ N(0, HHt)
## Measurement equation:
## y[t] = alpha[t] + eps[t], eps[t] ~ N(0, GGt)
y <- treering
y[c(3, 10)] <- NA # NA values can be handled
## Set constant parameters:
dt <- ct <- matrix(0)
Zt <- Tt <- array(1,c(1,1,1))
a0 <- y[1] # Estimation of the first width
P0 <- matrix(100) # Variance of 'a0'
## Estimate parameters:
fit.fkf <- optim(c(HHt = var(y, na.rm = TRUE) * .5,
GGt = var(y, na.rm = TRUE) * .5),
fn = function(par, ...)
-fkf(HHt = array(par[1],c(1,1,1)), GGt = array(par[2],c(1,1,1)), ...)$logLik,
yt = rbind(y), a0 = a0, P0 = P0, dt = dt, ct = ct,
Zt = Zt, Tt = Tt)
## Filter tree ring data with estimated parameters:
fkf.obj <- fkf(a0, P0, dt, ct, Tt, Zt, HHt = array(fit.fkf$par[1],c(1,1,1)),
GGt = array(fit.fkf$par[2],c(1,1,1)), yt = rbind(y))
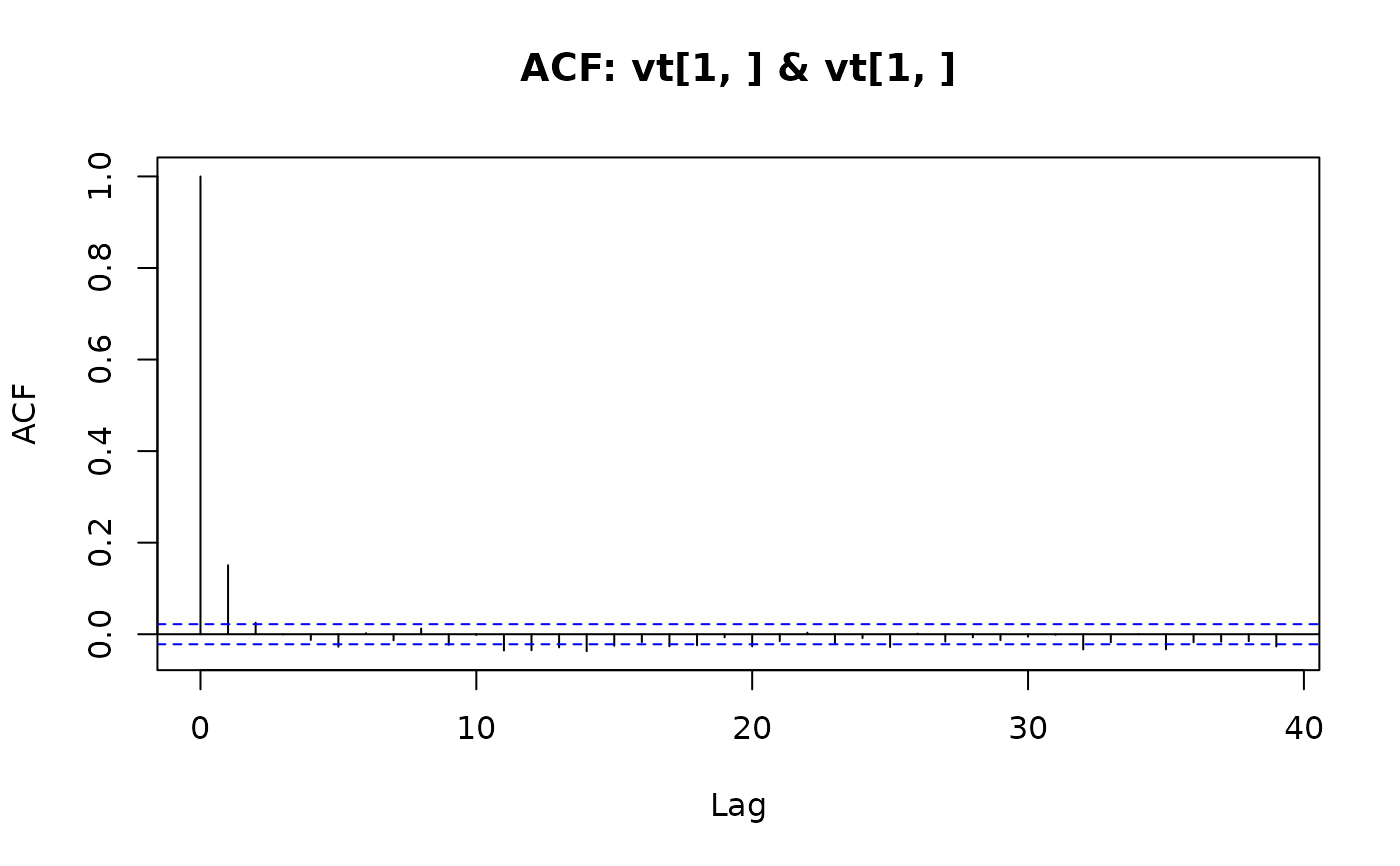
## Plot the width together with fitted local levels:
plot(y, main = "Treering data")
lines(ts(fkf.obj$att[1, ], start = start(y), frequency = frequency(y)), col = "blue")
legend("top", c("Treering data", "Local level"), col = c("black", "blue"), lty = 1)
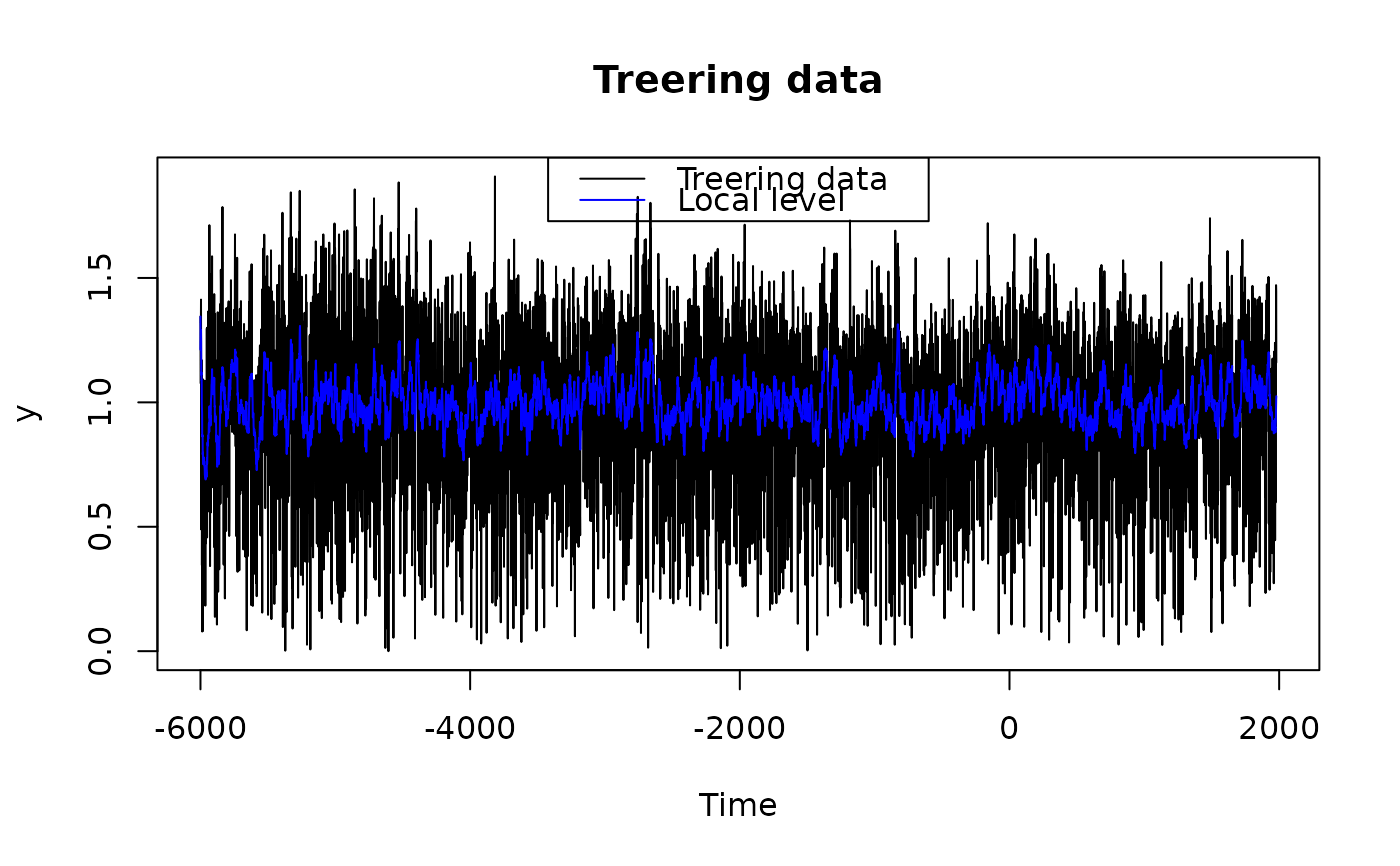 ## Check the residuals for normality:
plot(fkf.obj, type = "resid.qq")
## Check the residuals for normality:
plot(fkf.obj, type = "resid.qq")
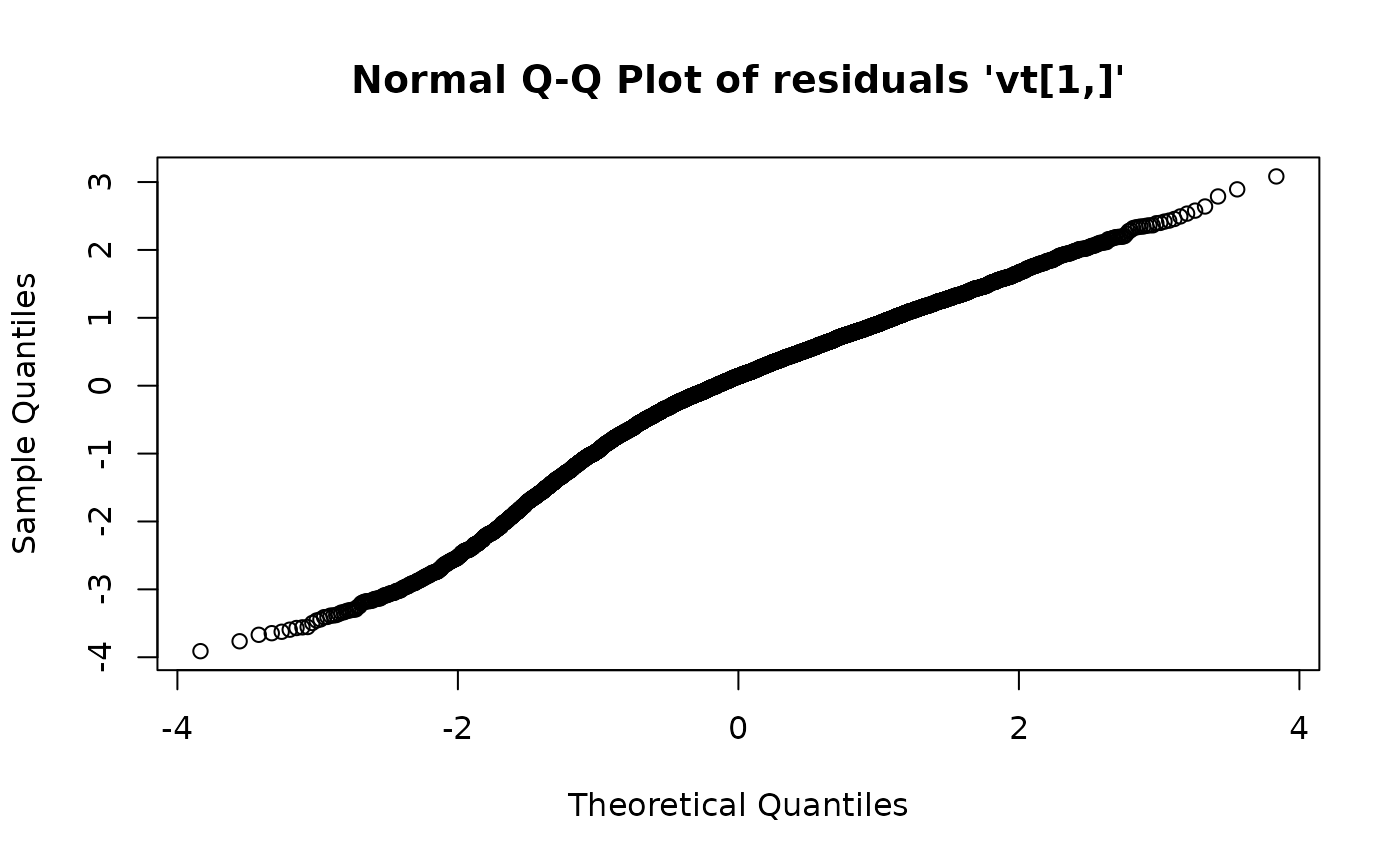 ## Test for autocorrelation:
plot(fkf.obj, type = "acf", na.action = na.pass)
## Test for autocorrelation:
plot(fkf.obj, type = "acf", na.action = na.pass)